Research
Research Overview
Identifying and understanding of variants of unknown significance (VUS) and development of resistance to targeted therapies represent two significant challenges facing current cancer care. Increased targeted panel sequencing of patients will continue to reveal variants lacking functional annotation. Thus, our lab strives to establish systematic methods to proactively assess the functionality of unknown variants in cancer. The second focus of our lab is to investigate the mechanisms underlying both on- and off-target sensitivity and resistance to targeted therapies. To address these important questions, we use a combination of high-throughput genetic screening, assay development, molecular biology, biochemistry, structural biology, and inhibitor sensitivity/resistance analyses in an effort to understand variant biology and improve clinical outcomes.
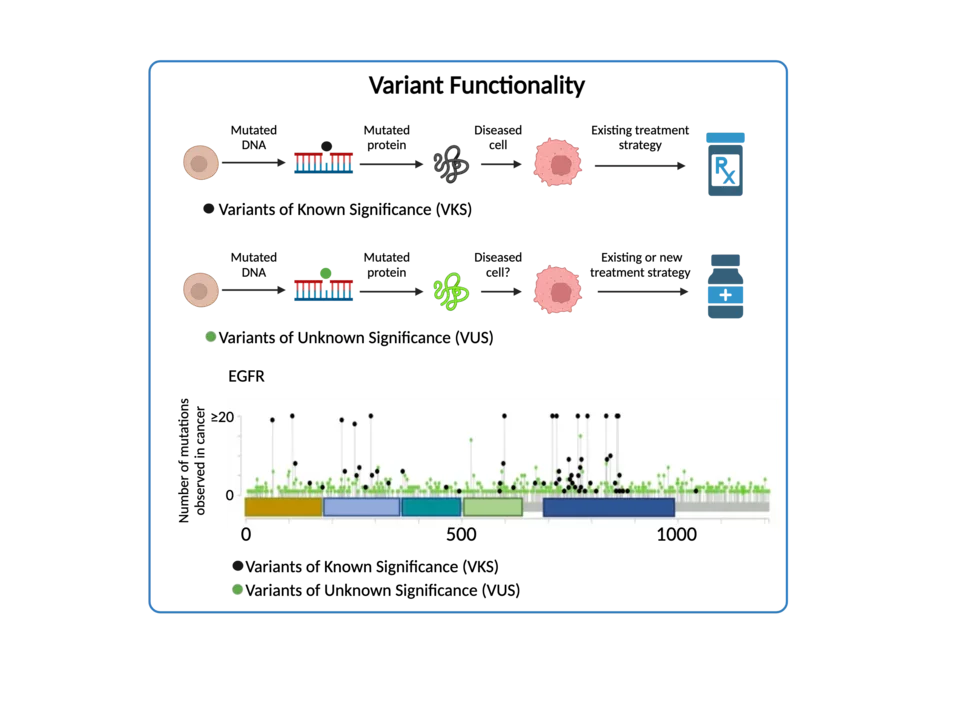
Variant Functionality
Variants of unknown significance (VUS) pose a significant challenge to global cancer care. VUS can be identified using sequencing technologies such as whole-genome or whole-exome sequencing, which analyze an individual's entire genetic code (WGS) or specific protein-coding regions (WES). During sequencing, genetic variations or variants, are detected by comparing an individual's DNA sequence to a reference genome. Variants that differ from the reference sequence are further analyzed to determine their potential significance. Those lacking clear annotation as either benign or pathogenic are classified as VUS. Understanding these variants is critical as they may underlie various genetic disorders, impacting patient health. Additionally, investigating uncharacterized variants can reveal novel disease mechanisms and therapeutic targets, which is essential for advancing cancer treatment. With personalized medicine on the rise, accurate interpretation of genetic variants, including VUS, is indispensable for tailoring treatments according to individual genetic profiles. Thus, investigations into VUS are fundamental for enhancing clinical care, driving genetic research, aiding informed decision-making in genetic counseling, and deepening our understanding of human genetics and disease.
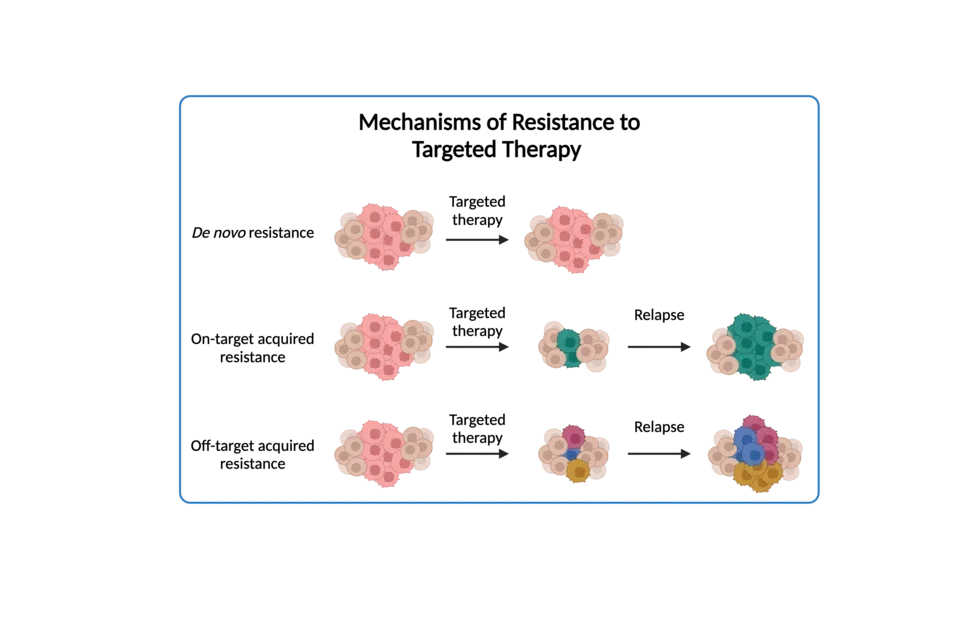
Resistance to targeted therapies
Despite initial success, many cancer patients eventually develop resistance to targeted therapies. Resistance mechanisms can arise through multiple pathways, including, but not limited to, genetic alterations, activation of bypass pathways, interactions with the tumor microenvironment, epigenetic modifications, and phenotypic heterogeneity. These mechanisms often lead to the reactivation of oncogenic pathways or the activation of alternative signaling pathways, rendering targeted therapies ineffective. Overcoming resistance to targeted therapies requires a multifaceted approach, including the development of combination therapies, novel targeted agents, and strategies to target resistance mechanisms directly. Understanding the underlying biology of resistance is crucial for improving treatment outcomes and developing more effective therapeutic strategies for cancer patients.